PUBLICATIONS
Selected publications
-

OriGene | A Self-Evolving Virtual Disease Biologist Automating Therapeutic Target Discovery
Therapeutic target discovery remains one of the most critical yet intuition-driven stages in drug development. We present OriGene, a self-evolving multi-agent system that functions as a virtual disease biologist to identify and prioritize therapeutic targets at scale. OriGene coordinates specialized agents that reason over diverse modalities, including genetic data, protein networks, pharmacological profiles, clinical records, and literature evidence, to generate and prioritize target discovery hypotheses. Through a self-evolving framework, OriGene continuously integrates human and experimental feedback to iteratively refine its core thinking templates, tool composition, and analytical protocols, thereby enhancing both accuracy and adaptability over time.
-
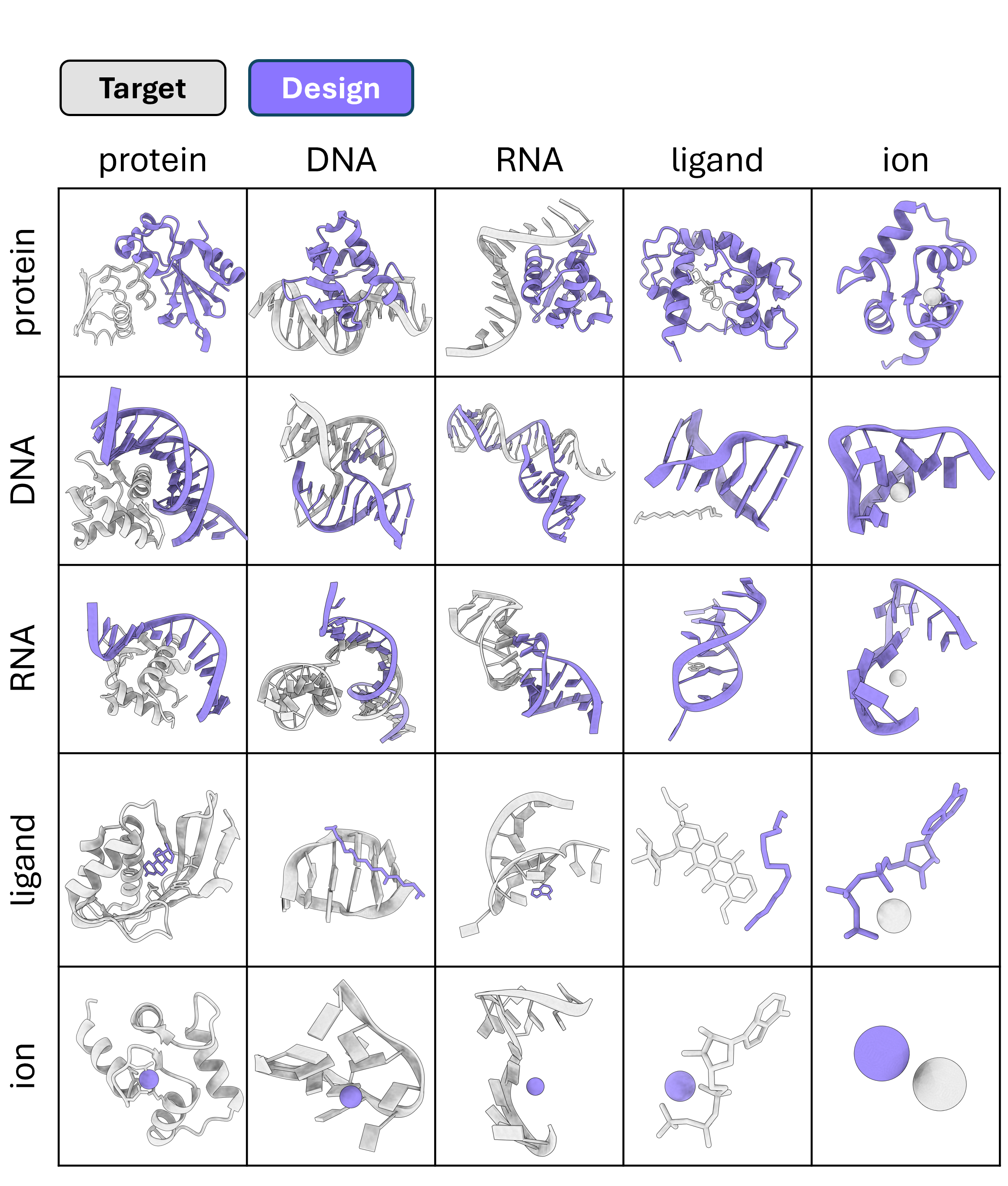
ODesign | A World Model for Biomolecular Interaction Design
Biomolecular interactions underpin almost all biological processes, and their rational design is central to programming new biological functions. Here we present ODesign, an all-atom generative world model for all-to-all biomolecular interaction design. ODesign allows scientists to specify epitopes on arbitrary targets and generate diverse classes of binding partners with fine-grained control. ODesign achieves state-of-the-art performance across protein, nucleic acid, and small-molecule design tasks, and has been experimentally validated by de novo designing pM-level binders against multiple targets.
Homepage: https://odesign.lglab.ac.cn/
-
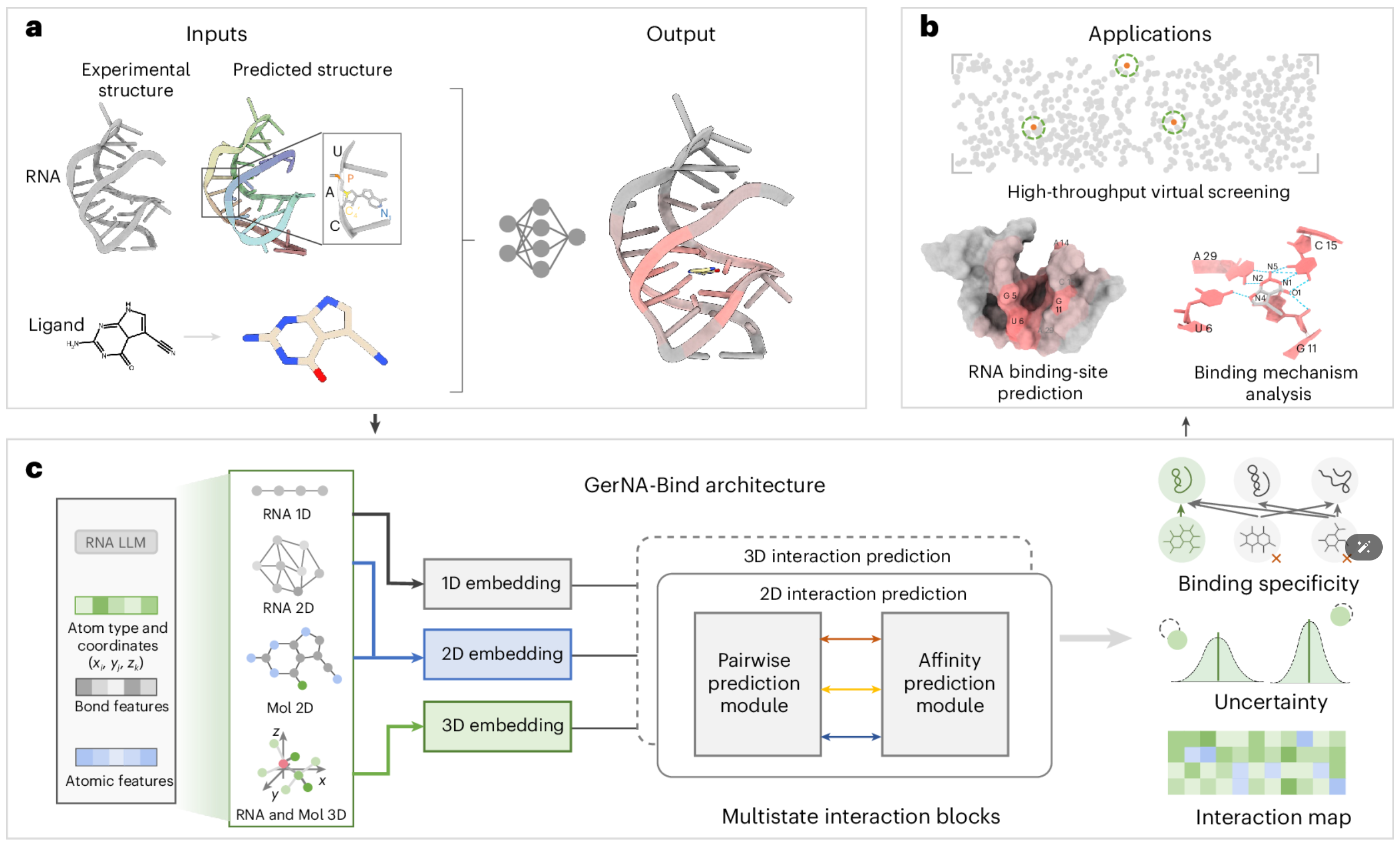
GerNA-Bind | Deciphering RNA–ligand binding specificity with GerNA-Bind
RNA molecules are essential regulators of biological processes and promising therapeutic targets for various diseases. Here we introduce GerNA-Bind, a geometric deep learning framework to predict RNA–ligand binding specificity by integrating multistate RNA–ligand representations and interactions. GerNA-Bind achieves state-of-the-art performance on multiple benchmark datasets and excels in predicting interactions for low-homology RNA–ligand pairs. In a large-scale virtual screening application, GerNA-Bind identified 18 structurally diverse compounds targeting the oncogenic MALAT1 RNA, with experimentally confirmed submicromolar affinities. Among them, one leading compound selectively binds the MALAT1 triple helix, reduces its transcript levels and inhibits cancer cell migration.
-
VCWorld | A Biological World Model for Virtual Cell Simulation
Virtual cell modeling aims to predict cellular responses to perturbations. We present VCWorld, a cell-level white-box simulator that integrates structured biological knowledge with the iterative reasoning capabilities of large language models to instantiate a biological world model. VCWorld operates in a data-efficient manner to reproduce perturbation-induced signaling cascades and generates interpretable, stepwise predictions alongside explicit mechanistic hypotheses. In drug perturbation benchmarks, VCWorld achieves state-of-the-art predictive performance, and the inferred mechanistic pathways are consistent with publicly available biological evidence.
-
DynamicBind | Predicting protein-ligand dynamic complex structure with generative models
Understanding protein function requires considering their dynamic nature, which traditional rigid docking methods overlook. Molecular dynamics simulations are accurate but computationally intensive. We introduce DynamicBind, a deep learning method using equivariant geometric diffusion networks to facilitate efficient transitions between protein equilibrium states. DynamicBind accurately predicts ligand-specific conformations from unbound protein structures without needing holo-structures, showing state-of-the-art performance in docking and virtual screening.
2025
Yong Liu, Chenqing Hua, Menglong Xu, Tao Zeng, Jiahua Rao, Zhongyue Zhang, Ruibo Wu, Jing-ke Weng, Connor W Coley, Shuangjia Zheng*. EnzymeCAGE: A Geometric Foundation Model for Enzyme Retrieval with Evolutionary Insights. Nature Catalysis, 2025. [Link][PDF]
Yunpeng Xia, Jiayi Li, Yi-Ting Chu, Jiahua Rao, Jing Chen, Chenqing Hua, Dong-Jun Yu, Xiu-Cai Chen*, Shuangjia Zheng*. GerNA-Bind: Geometric-enhanced RNA-ligand Binding Specificity Prediction with Deep Learning. Nature Machine Intelligence, 2025. [Link][PDF]
Odin Zhang, Jieyu Jin, Zhenxing Wu, Jintu Zhang, Po Yuan, Yuntao Yu, Haitao Lin, Haiyang Zhong, Xujun Zhang, Chenqing Hua, Weibo Zhao, Zhengshuo Zhang, Kejun Ying, Yufei Huang, Huifeng Zhao, Yu Kang, Peichen Pan, Jike Wang, Dong Guo, Shuangjia Zheng*, Chang-Yu Hsieh*, Tingjun Hou*. ECloudGen: Leveraging Electron Clouds as a Latent Variable to sScale Up Structure-based Molecular Design. Nature Computational Science, 2025. [Link][PDF]
Sheng Xu, Qiantai Feng, Lifeng Qiao, Hao Wu, Tao Shen, Yu Cheng, Shuangjia Zheng*, Siqi Sun*. FoldBench: An all-atom benchmark for biomolecular structure prediction. Nat. Commun., 2025. [Link][PDF]
Odin Zhang, Xujun Zhang, Haitao Lin, Cheng Tan, Qinghan Wang, Yuanle Mo, Qiantai Feng, Gang Du, Yuntao Yu, Zichang Jin, Ziyi You, Peicong Lin, Yijie Zhang, Yuyang Tao, Shicheng Chen, Jack Xiaoyu Chen, Chenqing Hua, Weibo Zhao, Runze Ma, Yunpeng Xia, Kejun Ying, Jun Li, Yundian Zeng, Lijun Lang, Peichen Pan, Hanqun Cao, Zihao Song, Bo Qiang, Jiaqi Wang, Pengfei Ji, Lei Bai, Jian Zhang, Chang-yu Hsieh, Pheng Ann Heng, Siqi Sun*, Tingjun Hou*, Shuangjia Zheng*. ODesign: A World Model for Biomolecular Interaction Design. arXiv, 2025. [Link][PDF]
Runze Ma, Zhongyue Zhang, Zichen Wang, Chenqing Hua, Zhuomin Zhou, Fenglei Cao, Jiahua Rao, Shuangjia Zheng*. RiboFlow: Conditional De Novo RNA Sequence-Structure Co-Design via Synergistic Flow Matching. NeurIPS,2025.[Link][PDF]
Dahao Xu, Jiahua Rao, Mingming Zhu, Jixian Zhang, Wei Lu, Shuangjia Zheng*, Yuedong Yang*. Accurately Predicting Protein Mutational Effects via a Hierarchical Many-Body Attention Network. NeurIPS, 2025.[Link][PDF]
Zhongyue Zhang, Jiahua Rao, Jie Zhong, Weiqiang Bai, Dongxue Wang, Shaobo Ning, Lifeng Qiao, Sheng Xu, Runze Ma, Will Hua, Jack Xiaoyu Chen, Odin Zhang, Wei Lu, Hanyi Feng, He Yang, Xinchao Shi, Rui Li, Wanli Ouyang, Xinzhu Ma, Jiahao Wang, Jixian Zhang, Jia Duan*, Siqi Sun*, Jian Zhang*, Shuangjia Zheng*. Fitness Aligned Structural Modeling enables Scalable Virtual Screening with AuroBind. arXiv, 2025. [Link][PDF]
Jiahao Wang, Shuangjia Zheng*. Efficient Protein Optimization via Structure-aware Hamiltonian Dynamics. AAAI, 2025.[Link][PDF]
Jiahua Rao, Hanjing Lin, Jiancong Xie, Zhen Wang, Shuangjia Zheng*, Yuedong Yang*. Incorporating Retrieval-based Causal Learning with Information Bottlenecks for Interpretable Molecular Graph Learning. KDD, 2025. [Link][PDF]
Qiujun Qiu, Shiyi Li, Jixian Zhang, Jixiang Chen, Xinyi Ding, Shengyao Liu, Jianyong Sheng, Zhiqing Pang, Ru Zhang, Anni Wang, Meichen Dong, Meng Zhang, Miaomiao Zhang, Tun Lu, Ning Gu, Shuigeng Zhou, Defang Ouyang, Dongsheng Li, Shuangjia Zheng*, Jianxin Wang*. Lab-in-the-loop Machine Learning for Brain-targeting Delivery System Design. Cell Biomaterials, 2025. [Link][PDF]
Jiaxuan Xia, Zicheng Gan, Jixian Zhang, Meichen Dong, Shengyao Liu, Bangchun Cui, Pengcheng Guo, Zhiqing Pang, Tun Lu, Ning Gu, Defang Ouyang, Chengtao Li, Shuangjia Zheng*, Jianxin Wang*. Geometric-aware Deep Learning enables Discovery of Bifunctional Ligand-based Liposomes for Tumor Targeting Therapy. Nano Today,2025.[Link][PDF]
Zhongyue Zhang, Zijie Qiu, Yingcheng Wu, Shuya Li, Dingyan Wang, Zhuomin Zhou, Duo An, Yuhan Chen, Yu Li, Yongbo Wang, Chubin Ou, Zichen Wang, Jack Xiaoyu Chen, Bo Zhang, Yusong Hu, Wenxin Zhang, Zhijian Wei, Runze Ma, Qingwu Liu, Bo Dong, Yuexi He, Qiantai Feng, Lei Bai*, Qiang Gao*, Siqi Sun*, Shuangjia Zheng*. OriGene: A Self-Evolving Virtual Disease Biologist Automating Therapeutic Target Discovery. BioRxiv, 2025. [Link][PDF]
Jiahua Rao, Hanjing Lin, Leyu Chen, Jiancong Xie, Shuangjia Zheng, Yuedong Yang. Multi-modal Contrastive Learning with Negative Sampling Calibration for Phenotypic Drug Discovery. CVPR, 2025. [Link][PDF]
Zichen Wang, Yaokun Ji, Jianing Tian, Shuangjia Zheng*. Retrieval Augmented Diffusion Model for Structure-informed Antibody Design and Optimization. ICLR, 2025. [Link][PDF]
Nianzu Yang, Songlin Jiang, Jian Ma, Huaijin Wu, Shuangjia Zheng, Wengong Jin, Junchi Yan. Repurposing alphafold3-like protein folding models for antibody sequence and structure co-design. NeurIPS,2025.[Link][PDF]
2024
Felix Wong, Dongchen He, Aarti Krishnan, Liang Hong, Alexander Z Wang, Jiuming Wang, Zhihang Hu, Satotaka Omori, Alicia Li, Jiahua Rao, Qinze Yu, Wengong Jin, Tianqing Zhang, Katherine Ilia, Jack X Chen, Shuangjia Zheng, Irwin King, Yu Li, James J Collins. Deep Generative Design of RNA Aptamers using Structural Predictions. Nature Computational Science, 2024. [Link][PDF]
Mei Li, Xiaoguang Liu, Hua Ji, Shuangjia Zheng*, Causal Subgraph Learning for Generalizable Inductive Relation Prediction. KDD, 2024.[Link][PDF]
Chenqing Hua, Bozitao Zhong, Sitao Luan, Liang Hong, Guy Wolf, Doina Precup, Shuangjia Zheng*, Reactzyme: A Benchmark for Enzyme-Reaction Prediction. NeurIPS, 2024.[Link][PDF]
Xiaoyi Liu, Chengwei Ai, Hongpeng Yang, Ruihan Dong, Jijun Tang, Shuangjia Zheng*, Fei Guo*. RetroCaptioner: beyond attention in end-to-end retrosynthesis transformer via contrastively captioned learnable graph representation. Bioinformatics, 2024.[Link][PDF]
Shuangjia Zheng*, Jiahua Rao, Jixian Zhang, Lianyu Zhou, Jiancong Xie, Ethan Cohen, Wei Lu, Chengtao Li, Yuedong Yang. Cross‐Modal Graph Contrastive Learning with Cellular Images. Advanced Science, 2024.[Link][PDF]
Tao Zeng, Zhehao Jin, Shuangjia Zheng, Tao Yu, Ruibo Wu, Developing BioNavi for Hybrid Retrosynthesis Planning. JACS Au, 2024.[Link][PDF]
Jiahua Rao, Jiancong Xie, Qianmu Yuan, Deqin Liu, Zhen Wang, Yutong Lu, Shuangjia Zheng*, Yuedong Yang*, A variational expectation-maximization framework for balanced multi-scale learning of protein and drug interactions. Nature Communications, 2024.[Link][PDF]
Jiancong Xie, Yi Wang, Jiahua Rao, Shuangjia Zheng*, Yuedong Yang*, Self-Supervised Contrastive Molecular Representation Learning with a Chemical Synthesis Knowledge Graph. Journal of Chemical Information and Modeling, 2024. [Link][PDF]
Chenqing Hua, Connor Coley, Guy Wolf, Doina Precup, Shuangjia Zheng*, Effective Protein-Protein Interaction Exploration with PPIretrieval. arXiv, 2024.[Link][PDF]
Wei Lu, Jixian Zhang, Weifeng Huang, Ziqiao Zhang, Xiangyu Jia, Zhenyu Wang, Leilei Shi, Chengtao Li, Peter G Wolynes, Shuangjia Zheng*, DynamicBind: predicting ligand-specific protein-ligand complex structure with a deep equivariant generative model. Nature Communications, 2024.[Link][PDF]
Wei Lu, Jixian Zhang, Jihua Rao, Zhongyue Zhang, Shuangjia Zheng*. AlphaFold3, a secret sauce for predicting mutational effects on protein-protein interactions. bioRxiv, 2024.[Link][PDF]
Before 2023
Jiahua Rao, Chengtao Li, Jixian Zhang, Yuedong Yang, Lianyu Zhou, Ethan Cohen, Wei Lu, Shuangjia Zheng,Cross-modal Graph Contrastive Learning with Cellular Images. Advanced Science, 2022.[Link][PDF]
Deqin Liu, Sheng Chen, Shuangjia Zheng, Sen Zhang, Yuedong Yang. SE (3) Equivalent Graph Attention Network as an Energy-Based Model for Protein Side Chain Conformation. in 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2023. IEEE.[Link][PDF]
Jiahua Rao, Shuangjia Zheng, Yuedong Yang, Integrating supercomputing and artificial intelligence for life science. Patterns, 2022.[Link][PDF]
Jiahua Rao, Shuangjia Zheng, Yutong Lu, Yuedong Yang,Quantitative evaluation of explainable graph neural networks for molecular property prediction. Patterns, 2022.[Link][PDF]
Youhai Tan, Lingxue Dai, Weifeng Huang, Yinfeng Guo, Shuangjia Zheng, Jinping Lei, Hongming Chen, Yuedong Yang,Drlinker: Deep reinforcement learning for optimization in fragment linking design. Journal of Chemical Information and Modeling, 2022. [Link][PDF]
Yi Wang, Shuangjia Zheng, Jiahua Rao, Yunan Luo, Yuedong Yang, ReaKE: Contrastive Molecular Representation Learning with Chemical Synthetic Knowledge Graph. Journal of Chemical Information and Modeling, 2022.[Link][PDF]
Shuangjia Zheng, Youhai Tan, Zhenyu Wang, Chengtao Li, Zhiqing Zhang, Xu Sang, Hongming Chen, Yuedong Yang, Accelerated rational PROTAC design via deep learning and molecular simulations. Nature Machine Intelligence, 2022.[Link][PDF]
Sijie Mai, Shuangjia Zheng, Ya Sun, Ying Zeng, Yuedong Yang, Haifeng Hu, Dynamic graph dropout for subgraph-based relation prediction. Knowledge-Based Systems, 2022.[Link][PDF]
Shuangjia Zheng, Tao Zeng, Chengtao Li, Binghong Chen, Connor W Coley, Yuedong Yang, Ruibo Wu, Deep learning driven biosynthetic pathways navigation for natural products with BioNavi-NP. Nature Communications, 2022.[Link][PDF]
Jiahua Rao, Shuangjia Zheng, Sijie Mai, Yuedong Yang, Communicative subgraph representation learning for multi-relational inductive drug-gene interaction prediction. IJCAI 2022.[Link][PDF]
Sijie Mai, Ying Zeng, Shuangjia Zheng, Haifeng Hu, Hybrid contrastive learning of tri-modal representation for multimodal sentiment analysis. IEEE Transactions on Affective Computing, 2022.[Link][PDF]
Qianmu Yuan, Sheng Chen, Jiahua Rao, Shuangjia Zheng, Huiying Zhao, Yuedong Yang, AlphaFold2-aware protein–DNA binding site prediction using graph transformer. Briefings in bioinformatics, 2022.[Link][PDF]
Penglei Wang, Shuangjia Zheng, Yize Jiang, Chengtao Li, Junhong Liu, Chang Wen, Atanas Patronov, Dahong Qian, Hongming Chen, Yuedong Yang, Structure-aware multimodal deep learning for drug–protein interaction prediction. Journal of chemical information and modeling, 2022.[Link][PDF]
Wei Lu, Qifeng Wu, Jixian Zhang, Jiahua Rao, Chengtao Li, Shuangjia Zheng, Tankbind: Trigonometry-aware neural networks for drug-protein binding structure prediction. Advances in neural information processing systems, 2022.[Link][PDF]
Xie, C., X.-X. Zhuang, Z. Niu, R. Ai, S. Lautrup, S. Zheng, Y. Jiang, R. Han, T.S. Gupta, and S. Cao, Amelioration of Alzheimer’s disease pathology by mitophagy inducers identified via machine learning and a cross-species workflow. Nature Biomedical Engineering, 2022.[Link][PDF]
Jianzhao Gao, Shuangjia Zheng, Mengting Yao, Peikun Wu, Precise estimation of residue relative solvent accessible area from Cα atom distance matrix using a deep learning method. Bioinformatics, 2022.[Link][PDF]
Shuangjia Zheng, Ying Song, Zhang Pan, Chengtao Li, Le Song, Yuedong Yang, Molecular Attributes Transfer from Non-Parallel Data. arXiv preprint 2021.[Link]
Lizhao Hu, Yuyao Yang, Shuangjia Zheng, Jun Xu, Ting Ran, Hongming Chen, Kinase inhibitor scaffold hopping with deep learning approaches. Journal of Chemical Information and Modeling, 2021.[Link]
Shuangjia Zheng, Zengrong Lei, Haitao Ai, Hongming Chen, Daiguo Deng, Yuedong Yang, Deep scaffold hopping with multimodal transformer neural networks. Journal of cheminformatics, 2021.[Link]
Shuangjia Zheng, Sijie Mai, Ya Sun, Haifeng Hu, Yuedong Yang, Subgraph-aware few-shot inductive link prediction via meta-learning. IEEE Transactions on Knowledge and Data Engineering, 2022.[Link]
Jianwen Chen✝, Shuangjia Zheng✝, Ying Song, Jiahua Rao, Yuedong Yang, Learning attributed graph representations with communicative message passing transformer. IJCAI 2021.[Link]
Shuangjia Zheng, Jiahua Rao, Ying Song, Jixian Zhang, Xianglu Xiao, Evandro Fei Fang, Yuedong Yang, Zhangming Niu, PharmKG: a dedicated knowledge graph benchmark for bomedical data mining. Briefings in bioinformatics, 2021.[Link]
Shuangjia Zheng, Tao Zeng, Chengtao Li, Binghong Chen, Connor W Coley, Yuedong Yang, Ruibo Wu, BioNavi-NP: Biosynthesis Navigator for Natural Products. Nature Communications 2021.[Link]
Jiahao Wang, Shuangjia Zheng, Jianwen Chen, Yuedong Yang, Meta learning for low-resource molecular optimization. Journal of Chemical Information and Modeling, 2021.[Link]
Ying Song*, Shuangjia Zheng*, Liang Li, Xiang Zhang, Xiaodong Zhang, Ziwang Huang, Jianwen Chen, Huiying Zhao, Yusheng Jie, Ruixuan Wang, Yutian Chong, Jun Shen, Yunfei Zha, Yuedong Yang, Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM transactions on computational biology and bioinformatics, 2021.[Link]
Jiahua Rao, Shuangjia Zheng, Ying Song, Jianwen Chen, Chengtao Li, Jiancong Xie, Hui Yang, Hongming Chen, Yuedong Yang, MolRep: A deep representation learning library for molecular property prediction. bioRxiv, 2021.[Link]
Zhihong Liu, Dane Huang, Shuangjia Zheng, Ying Song, Bingdong Liu, Jingyuan Sun, Zhangming Niu, Qiong Gu, Jun Xu, Liwei Xie, Deep learning enables discovery of highly potent anti-osteoporosis natural products. European Journal of Medicinal Chemistry, 2021.[Link]
Pan Zhang, Shuangjia Zheng, Jianwen Chen, Yaoqi Zhou, Yuedong Yang. DeepANIS: Predicting antibody paratope from concatenated CDR sequences by integrating bidirectional long-short-term memory and transformer neural networks. in 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2021. IEEE.[Link]
Sijie Mai*, Shuangjia Zheng*, Yuedong Yang, Haifeng Hu. Communicative message passing for inductive relation reasoning. in Proceedings of the AAAI Conference on Artificial Intelligence. 2021.[Link]
Shimin Su, Yuyao Yang, Hanlin Gan, Shuangjia Zheng, Fenglong Gu, Chao Zhao, Jun Xu, Predicting the feasibility of copper (i)-catalyzed alkyne–azide cycloaddition reactions using a recurrent neural network with a self-attention mechanism. Journal of Chemical Information and Modeling, 2020.[Link]
Shuangjia Zheng, Yongjian Li, Sheng Chen, Jun Xu, Yuedong Yang, Predicting drug–protein interaction using quasi-visual question answering system. Nature Machine Intelligence, 2020.[Link]
Chaochao Yan, Qianggang Ding, Peilin Zhao, Shuangjia Zheng, Jinyu Yang, Yang Yu, Junzhou Huang, Retroxpert: Decompose retrosynthesis prediction like a chemist. Advances in Neural Information Processing Systems, 2020.[Link]
Ying Song*, Shuangjia Zheng*, Zhangming Niu, Zhang-Hua Fu, Yutong Lu, Yuedong Yang. Communicative Representation Learning on Attributed Molecular Graphs. IJCAI. 2020.[Link]
Jianwen Chen*, Shuangjia Zheng*, Huiying Zhao, Yuedong Yang, Structure-aware protein solubility prediction from sequence through graph convolutional network and predicted contact map. Journal of cheminformatics, 2021.[Link]
Yuyao Yang*, Shuangjia Zheng*, Shimin Su, Chao Zhao, Jun Xu, Hongming Chen, SyntaLinker: automatic fragment linking with deep conditional transformer neural networks. Chemical science, 2020.[Link]
Zhe Sun, Shuangjia Zheng, Huiying Zhao, Zhangming Niu, Yutong Lu, Yi Pan, Yuedong Yang, To improve prediction of binding residues with DNA, RNA, carbohydrate, and peptide via multi-task deep neural networks. IEEE/ACM transactions on computational biology and bioinformatics, 2021.[Link]
Shuangjia Zheng, Jiahua Rao, Zhongyue Zhang, Jun Xu, Yuedong Yang, Predicting retrosynthetic reactions using self-corrected transformer neural networks. Journal of chemical information and modeling, 2019.[Link]
Shuangjia Zheng, Xin Yan, Qiong Gu, Yuedong Yang, Yunfei Du, Yutong Lu, Jun Xu, QBMG: quasi-biogenic molecule generator with deep recurrent neural network. Journal of cheminformatics, 2019.[Link]
Shuangjia Zheng, Xin Yan, Yuedong Yang, Jun Xu, Identifying structure–property relationships through smiles syntax analysis with self-attention mechanism. Journal of chemical information and modeling, 2019.[Link]

