METHODS
-
 DynamicBind is a deep learning method using equivariant geometric diffusion networks to facilitate efficient transitions between protein equilibrium states. It recovers ligand-specific conformations from unbound protein structures promoting efficient transitions between different equilibrium states.
DynamicBind is a deep learning method using equivariant geometric diffusion networks to facilitate efficient transitions between protein equilibrium states. It recovers ligand-specific conformations from unbound protein structures promoting efficient transitions between different equilibrium states. Github Fork 50+Github Star 220+
Github Fork 50+Github Star 220+ -
 TankBind is a geometric deep learning method for molecular rigid docking. It could predict both the protein-ligand binding structure and their affinity. And it also support virtual screening.
TankBind is a geometric deep learning method for molecular rigid docking. It could predict both the protein-ligand binding structure and their affinity. And it also support virtual screening. Github Fork 40+Github Star 180+
Github Fork 40+Github Star 180+ -
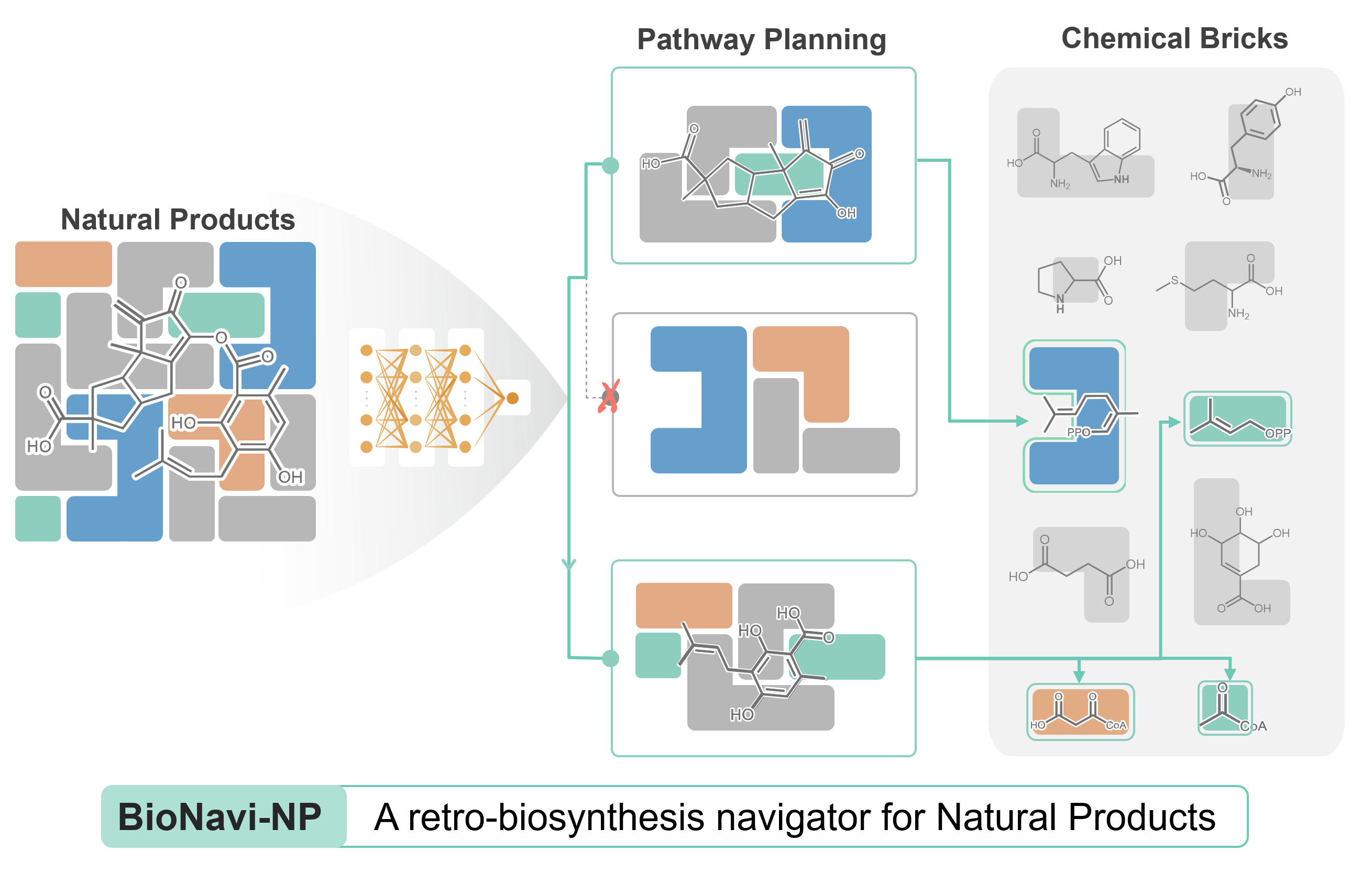 BioNavi-NP is a user-friendly toolkit that predicts biosynthetic pathways for NPs and NP-like compounds.The toolkit as well as the curated datasets and learned models are freely available to facilitate the elucidation and reconstruction of the biosynthetic pathways for NPs.
BioNavi-NP is a user-friendly toolkit that predicts biosynthetic pathways for NPs and NP-like compounds.The toolkit as well as the curated datasets and learned models are freely available to facilitate the elucidation and reconstruction of the biosynthetic pathways for NPs. Github Fork 14Github Star 45+
Github Fork 14Github Star 45+ -
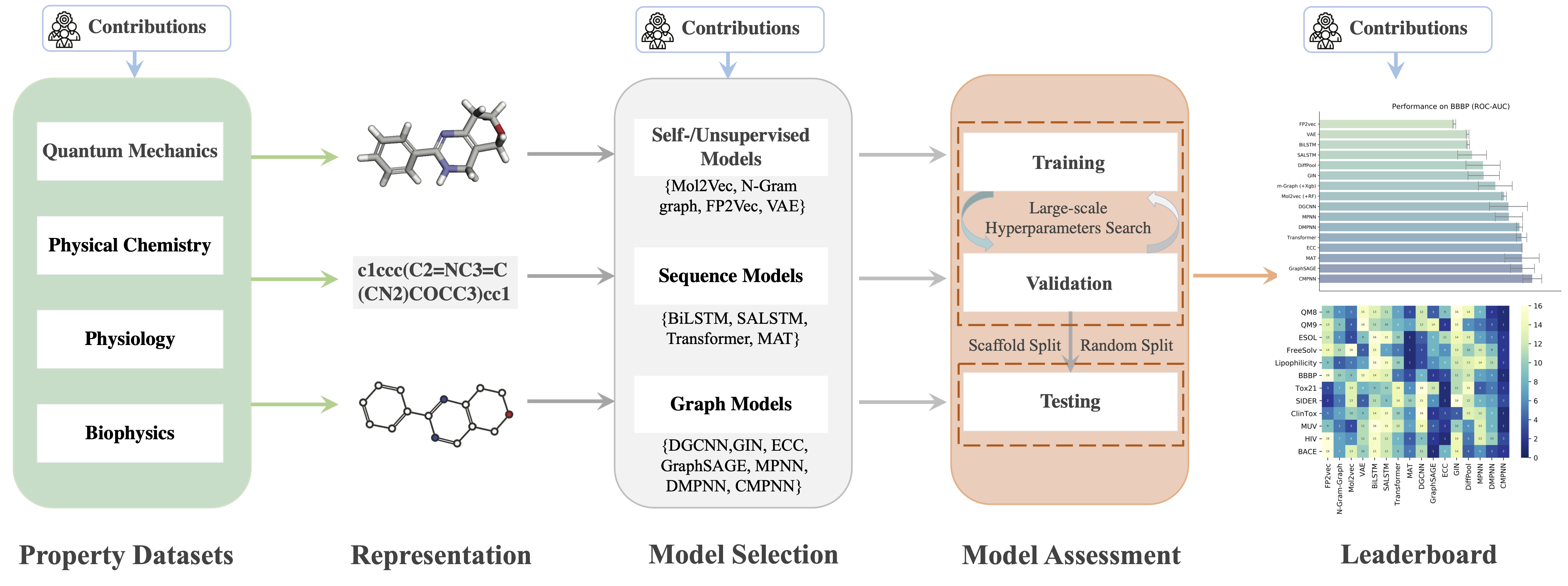 MolRep is a Python package for fairly measuring algorithmic progress on chemical property datasets. It currently provides a complete re-evaluation of 16 state-of-the-art deep representation models over 16 benchmark property datsaets.
MolRep is a Python package for fairly measuring algorithmic progress on chemical property datasets. It currently provides a complete re-evaluation of 16 state-of-the-art deep representation models over 16 benchmark property datsaets. Github Fork 30+Github Star 120+
Github Fork 30+Github Star 120+ -
 DrugVQA is a multimodel learning method for protein-ligand interaction prediction.
DrugVQA is a multimodel learning method for protein-ligand interaction prediction. Github Fork 30+Github Star 120+
Github Fork 30+Github Star 120+

